Data Interpretation and Discovery Partner for Inherited Disease
InheriNext®, a platform by Compass Bioinformatics, has your NGS analysis needs at heart.
With its cutting-edge variant ranking algorithm, intuitive design & validated accuracy, we empower scientists and clinicians in hospitals and research laboratories to drive discovery and diagnosis.

InheriNext® transparency delivers
Increases Trust with Confidence
Knowing how an algorithm reaches its conclusions is essential. InheriNext® reveals all supporting evidence—from triggered ACMG rules to ClinVar examples that align with or challenge the findings.
Improves Understanding
Linked sources facilitate a deeper understanding of InheriNext®. You can assess the reliability and applicability of the results for individual cases.
Enhances Clinical Judgment
You aren't a passive recipient of algorithm outputs! InheriNext® leaves the ability to critically evaluate the results and incorporate your clinical judgment into the decision-making process. Customizable ACMG options, filter settings, and reporting options empower your clinical judgement.
Reduces Bias
Understanding the algorithms allows you to identify potential biases or limitations that may affect the results. This promotes fairness and equity in healthcare delivery.
Facilitates Collaboration
Transparent algorithms and shareable databases allow your team to collaborate more effectively in interpreting and applying the results of individual cases or patient cohorts.
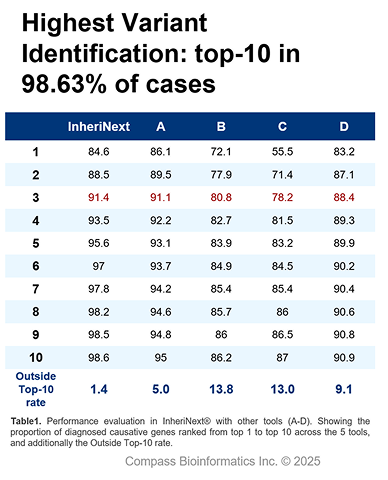
InheriNext® helps researchers and clinicians uncover the genetic causes behind inherited diseases
Sequencing → → →
InheriNext® supports data from most automated sequencing instruments. Contact us if you would like to test the compatibility of your data!
Secondary Analysis

Utilize secure & optimized pipelines for complex variant calling from FASTQ or VCF formats.
Interpretation

Identify & rank causative variants based on clinical phenotypes, in silico gene panels, or customized filters!
Reporting

Custom report functionality for inherited diseases including summaries & support for categorized variants.
Why InheriNext® ?
InheriNext® combines cutting-edge AI with user-friendly tools to simplify the complexities of NGS genetic testing. Leverage our expertise to manage massive datasets, uncover meaningful insights, and streamline your workflows with intuitive solutions.
Whether you interpret cases one at a time, in families or in cohorts, InheriNext® automates routine tasks with accuracy, efficiency and reliability, so you can focus on advancing patient care.
5-15 min
Prioritize variants quickly and efficiently—turnaround times from VCF upload to ranked variants are as fast as 3 minutes for WES & 15 minutes for WGS.
Transparency
One click in the results table reveals the data behind each variant score and classification.
Best Practice
Automation means consistency. Baked-in ACMG guidelines ensure confidence.
Easy Validation
Easily cross-check results with color-coded scores from prediction models, databases & intuitive infographics!
Margin Sensitive Pricing
Optimized for large-scale genetic diagnoses, keeping costs manageable.
Core Product Modules
Easily manage patient data, generate custom reports, and build your own databases — all through an intuitive interface designed to simplify your workflow.
Workflow Runner

Automates the conversion of raw sequencing data into actionable insights, streamlining NGS workflows for faster, accurate genetic analysis.
Phenotype Gene Prioritizer

Ranks & interprets causative variants based on phenotype & variant pathogenicity, ACMG guidelines, prediction models, and more!
PhenoVarDB

Expands the value of Individual cases in the context of cohorts from your lab or your community.
Testimonials
Join the growing network of professionals who save time and achieve higher diagnostic yield with InheriNext®!
InheriNext® began in Taiwan, where physicians from leading medical centers rely on our technology every day. Here's what our clients have to say:
"Your analysis software interface is very user-friendly — I support you fully!"

Ryan Chang Ph.D.,
Pharmatech R&D Manager
"InheriNext is a great system!"

Dr. Yung-Ting Kuo,
Pediatrics Dept. Director
"The Compass team is dedicated to innovation, and I am highly optimistic about its future in the WGS era!"

Dr. Yi-Chung Lee,
Attending Physician & Adjunct Prof. of Neurology
Partner With Us
Our product is already transforming the way genetic data is analyzed, but we know every organization has unique needs.
We’re looking for a strategic partners to collaborate with us as we fine-tune and enhance our solution. Together, we can tailor our platform to perfectly align with your goals while solving your most pressing challenges.
Interested in collaborating? Let us know:

Associated Publications
Eye (2024)
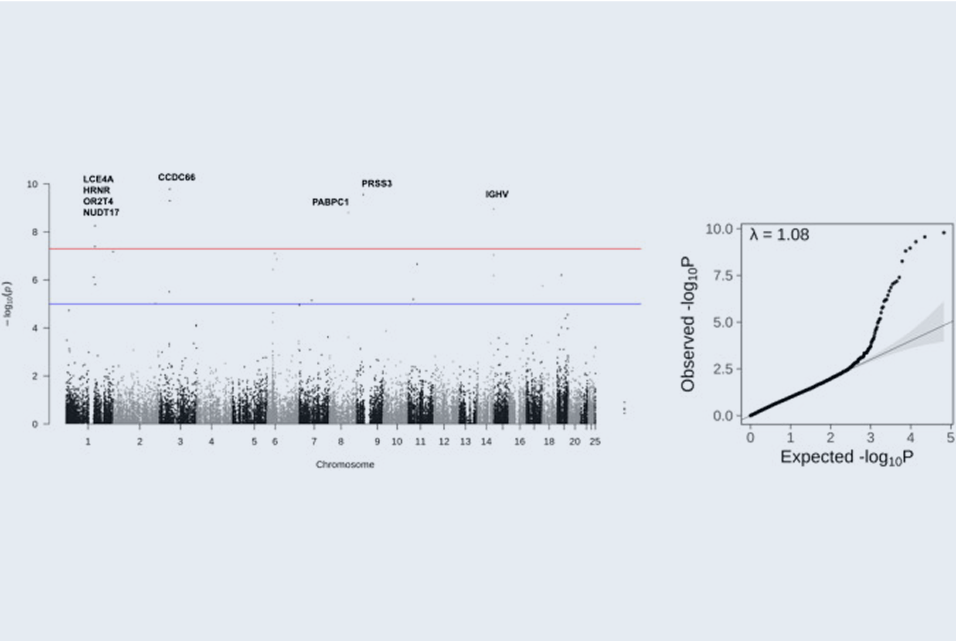
Conclusions: Several candidate susceptibility genes including RP1L1, RPGR, RPE65 and CCDC66 were identified to be associated with CQ/HCQ retinopathy. In addition to disease susceptibility, patients with increased genetic variants are more vulnerable to poor visual outcomes.
Annals of Clinical and Translational Neurology (2023)
Conclusions: Hereditary spastic paraplegia type 54 (SPG54) is a neurodegenerative disorder caused by DDHD2 gene mutations, leading to progressive lower limb spasticity and additional neurological symptoms. This study identified SPG54 in 1.2% of 242 HSP patients using mutational analysis of DDHD2, uncovering novel mutations that reduce DDHD2 protein levels, confirming their role in the disease.
International Journal of Molecular Sciences (2022)
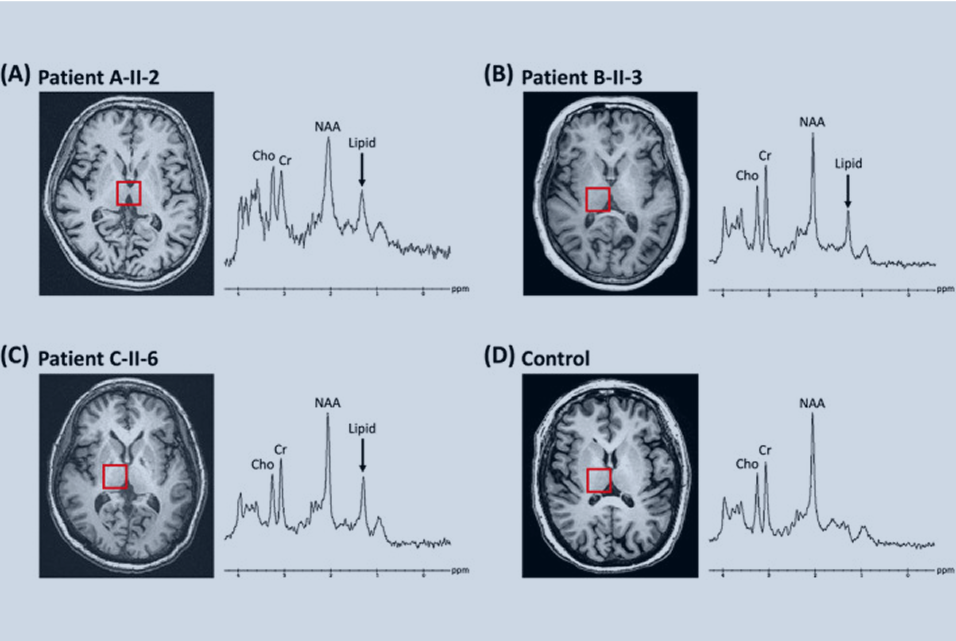
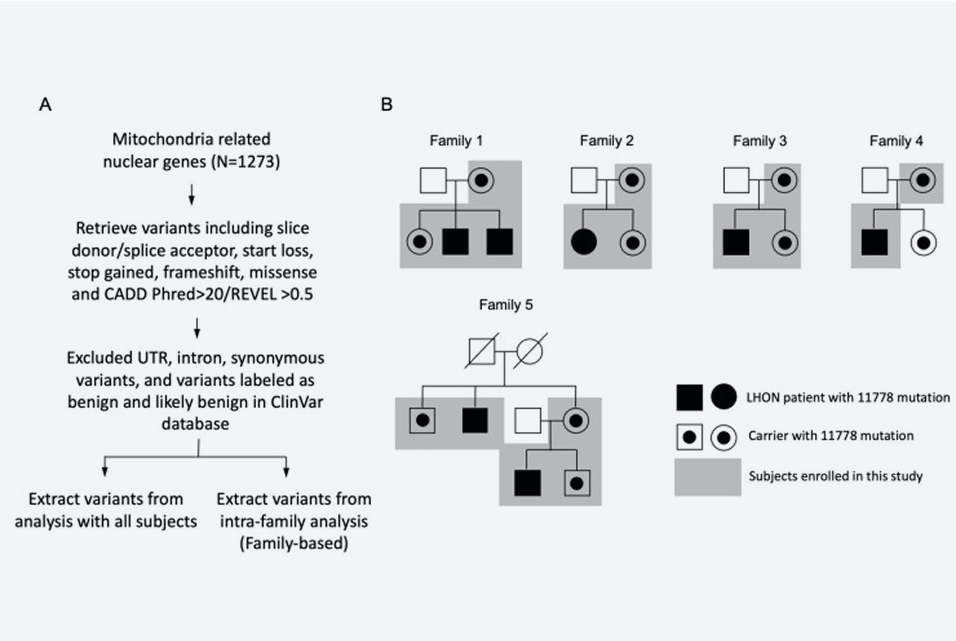
Conclusions: Family-based analysis showed that several candidate MT genes including METAP1D (c.41G > T), ACACB (c.1029del), ME3 (c.972G > C), NIPSNAP3B (c.280G > C, c.476C > G), and NSUN4 (c.4A > G) were involved in the penetrance of LHON.
Frequently Asked Questions
How can I test InheriNext® capabilities?
We offer a four-stage process designed to ensure InheriNext meets your specific needs and integrates seamlessly into your workflow: 1) a personal demonstration, 2) a trial, with sample data and your data. 3) a pilot license and 4) a custom integration into your operations.
Can I get a demo of InheriNext®?
Click the “Book a Demo” button at the top of the page, schedule a personalized demonstration between you and our scientists at Compass. In the comments, please provide information so we can tailor the presentation to your requirements and confirm data compatibility. We want to ensure that your time during the demonstration answers your questions and allows you to experience InheriNext® user-friendly interface and interpretation capabilities.
Can I get a trial of InheriNext®?
Prior to a trial, you should have a demonstration, to familiarize yourself with the platform. Following the demonstration, you’ll receive a trial account with access to either your own data or our sample datasets (VCF or FASTQ files, depending on your workflow). Providing aligned phenotypic data enhances the trial’s value. (During the process, we may request access to provide optimal support). Click the “Book a Demo” button to schedule a demonstration.
Can I get a pilot license for InheriNext®?
Following the trial period, we can provide a pilot license to test the integration of InheriNext® into your institution. Our staged pilot program includes additional training and allows for customization discussions to ensure a smooth transition. Click on “Contact Us” to find out more!
What does a contract for InheriNext® look like?
We offer transparent pricing models (annual licenses, per-report options, and collaborative partnerships) and are happy to discuss contract terms at any stage of the trial. Our goal is a mutually beneficial relationship that prioritizes patient care. Customization options include report generation, panel support for various sequencing protocols, and seamless IT integrations.